ABOVE: Astronaut Eytan Stibbe tested an RPA-CRISPR-Cas12a genetic detection assay on board the International Space Station. Rakia Mission
Cetus biochemist Kary Mullis’ invention of polymerase chain reaction (PCR) in 1985 revolutionized genetics and won him a share of the 1993 Nobel Prize in Chemistry.1 PCR was instrumental in the sequencing of the human genome, and today, it is used for everything from diagnosing genetic disorders to studying the evolutionary relationships between species.2,3
Despite its utility, PCR’s main limitation is that it requires precise cycles of heating and cooling to amplify DNA. The thermal cyclers that perform this operation are clunky, relatively expensive, and consume a lot of power, making them unsuitable for use in the field or in low-resource settings.
Just the warmth of your hand is enough to drive the reaction.
—Derek Stemple, Camena Bioscience
In 2006, researchers Olaf Piepenburg, Colin Williams, and Niall Armes of ASM Scientific, which later became TwistDx, along with Derek Stemple, at the time a molecular biologist at the Wellcome Trust Sanger Institute and currently the chief science officer at Camena Bioscience, developed an alternative. They invented recombinase polymerase amplification (RPA), an isothermal amplification technique that functions at relatively low temperatures.4 “Just the warmth of your hand is enough to drive the reaction,” said Stemple.
Since its invention, researchers have demonstrated a myriad of applications for this technology, including rapid identification of plant, animal, and human pathogens, including SARS-CoV-2, detecting antimicrobial resistance genes, and water quality monitoring.5–10
The beginnings of RPA
The researchers didn’t originally set out to develop a new isothermal amplification method, said Piepenburg. Instead, they were trying to develop a new technique for single molecule sequencing. During this project, they became interested in ways to sequence specific sites within the genome, rather than random pieces.
“Niall’s idea was to use recombinase proteins like RecA to target DNA primers to specific sites,” said Piepenburg. “This idea evolved into the notion that if you have two primers that are interacting with recombinase proteins and they move in opposite directions along a double-stranded piece of DNA, wouldn’t you start to get an amplification reaction?”
RecA is a bacterial protein that is crucial for repairing double-stranded breaks in DNA via the process of homologous recombination. RecA binds to single-stranded DNA, searches along a separate piece of double stranded DNA for the cognate sequence, and then promotes DNA strand invasion.11 After testing various proteins in the RecA family, the researchers ultimately settled on the phage protein uvsX recombinase, which performs a similar function. In RPA, this recombinase protein binds to primers designed by scientists, then scans the DNA, and inserts the primers into complementary sites in the DNA. Then single-stranded binding proteins called gp32 bind to the displaced DNA strand to stabilize it. The recombinase disassembles, allowing a DNA polymerase to bind to the 3’ end of the primer, and DNA synthesis begins.
“We tested a lot of different polymerases,” said Stemple. “The polymerases that worked the best were the ones that were able to displace the second strand, that don’t need single-stranded DNA to work from, like Taq polymerase.” The researchers eventually found a polymerase from Bacillus subtilis that was especially effective. “It’s like a cow catcher at the front of the train, separating double stranded DNA at the front of the polymerase,” said Stemple.
This technique enables rapid amplification of the target DNA: the process takes just 20 to 40 minutes.12 Once the target DNA has been amplified, researchers can use a variety of strategies to detect the DNA, including lateral flow and fluorescence-based tests.
The researchers also demonstrated the real-world utility of this technique. “We really wanted to show that we could detect disease-causing bacteria in very small numbers,” said Stemple. Indeed, by combining RPA with a fluorescent probe detection system, they detected just ten copies of DNA from methicillin-resistant Staphylococcus aureus (MRSA), a pathogen responsible for an estimated 100,000 deaths per year.13
“After we published the paper, we got an email from Kary Mullis congratulating us,” said Stemple. “That was pretty cool.”
RECOMBINASE POLYMERASE AMPLIFICATION IN ACTIONA rapid isothermal amplification technique enables pathogen identification and antibiotic resistance detection in low-resource settings. | |
Recombinase polymerase amplification (RPA) is a technique for rapidly copying segments of DNA. Unlike polymerase chain reaction (PCR), it does not require thermal cycling, so less equipment is needed. | |
HOW RPA WORKSResearchers require three defining elements for RPA: uvsX recombinase proteins, single-stranded binding proteins, and strand-displacing DNA polymerases. They also need primers and nucleotides to amplify the target DNA. |  |
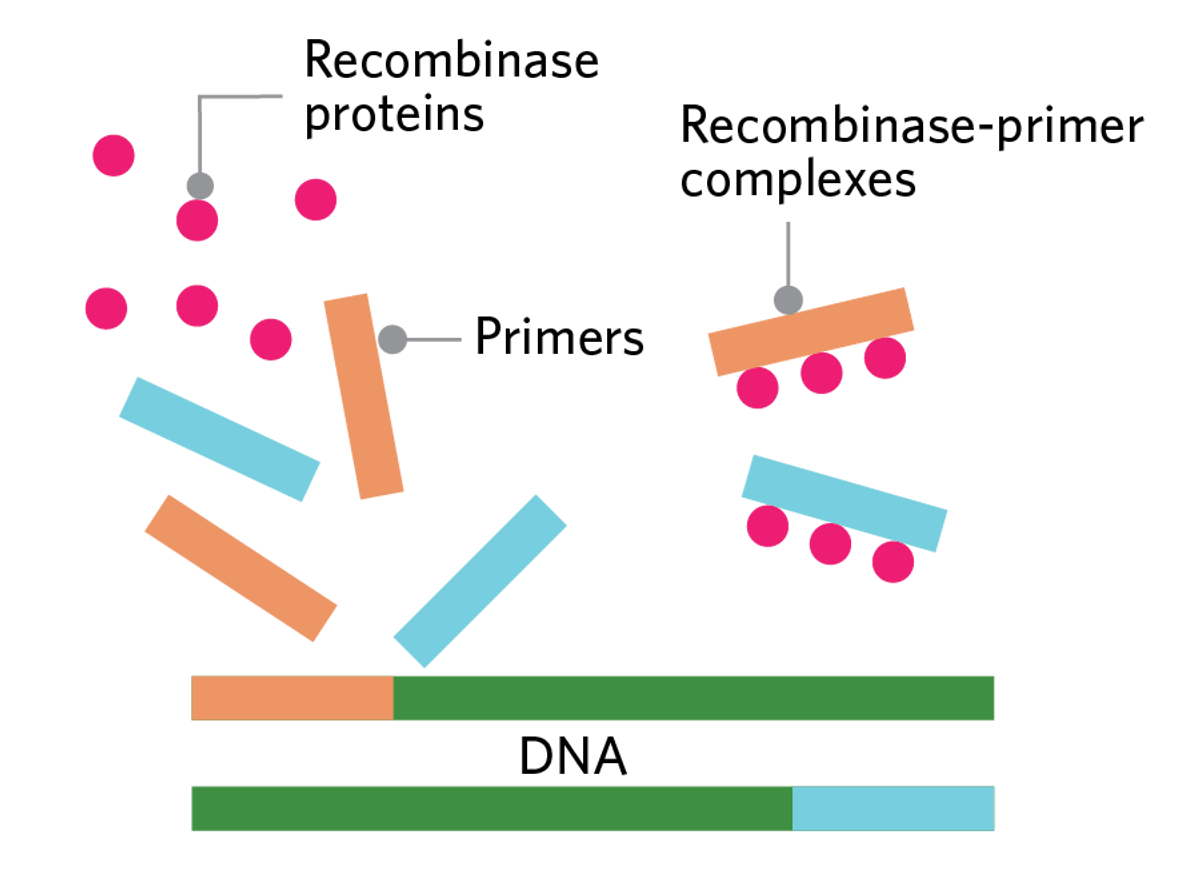 | First, the primers bind to the uvsX recombinase proteins to form recombinase-primer complexes. |
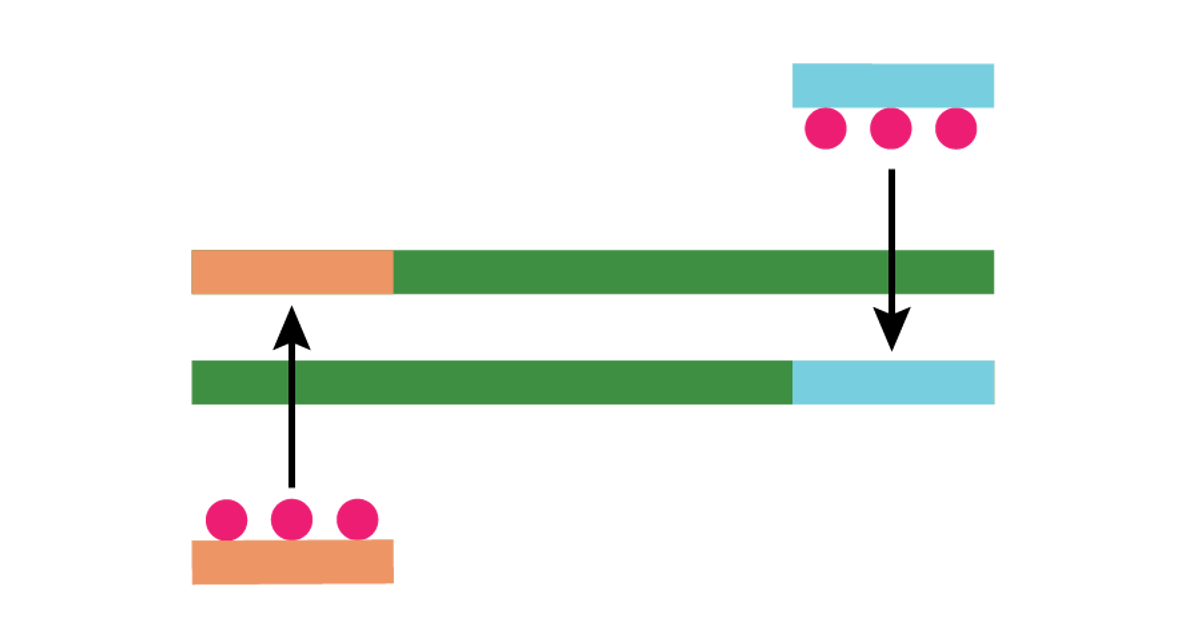 | Then the recombinase inserts the primers into complementary sites in the DNA. |
 | Single-stranded binding proteins bind to the displaced DNA strand and stabilize it. |
 | The recombinase disassembles and the strand-displacing DNA polymerase binds to the 3’ end of the primer. |
 | The polymerase elongates the primer. |
 | The target DNA strands are duplicated. |
Detecting DNA from the cervix to outer space
“A question that we faced early on when presenting at scientific conferences or talking to potential collaborators was, ‘Why not just use PCR?’” said Piepenburg. “PCR is excellent at what it does, but it is limited by the fact that you need a heavy instrument that takes a lot of energy to run. So, it’s very hard to see PCR ever reaching a stage where you can make a completely disposable diagnostic. And especially for rapid diagnostics, that’s really the holy grail.”
With RPA in their toolkit, researchers around the world can now develop rapid tests for various plant, animal, and human pathogens that can be used in places far afield from traditional laboratory settings, with important implications for both agriculture and human health.7,14,15
One such application is human papillomavirus (HPV) testing, which has long been difficult in low-resource settings. “There is a huge need for point-of-care technologies that can be used anywhere in the world; remote locations and developing countries are really in need of tests that can accurately detect HPV in women,” said Sylvia Daunert, a biochemist and molecular biologist at the University of Miami.
The test also needs to be fast. In these settings, said Daunert, “if women leave the community health clinic without being treated, the likelihood that they will come back is very low.”
“What makes RPA really good for this application is that it works at just one temperature, which is relatively low compared to other isothermal methods, some of which need to be kept at 65 degrees Celsius or require initial heating,” said Daunert.
Daunert and her team designed consensus primers that could amplify any of the 14 high-risk HPV genotypes. They lyophilized the reagents and combined them with primers into one tube, reducing the number of steps and eliminating the need for precise pipetting. “We wanted anyone to be able to operate this test,” said Daunert.
Their test detected high-risk HPV with a sensitivity of 96 percent and a specificity of 83 percent.7 Daunert’s next step will be testing the diagnostic on the ground in low-resource settings in regions like central Africa.
Some researchers deploy RPA even farther afield. Gur Pines, who studies molecular diagnostics and detection at the Agricultural Research Organization–Volcani Institute, was working on a rapid molecular detection strategy for the peach fruit fly, an important agricultural pest, when he got the opportunity to send an experiment to space as part of Israel’s Rakia Mission.
Pines studied a detection strategy that combined RPA and CRISPR technology. RPA amplifies the DNA, and when CRISPR-Cas12a binds to the target DNA, the Cas12a begins to indiscriminately chop up single-stranded DNA. (This nonspecific cutting of single-stranded DNA is unique to Cas12a and is not shared by the more well-known Cas9 protein.) Researchers use probes with a fluorophore and a quencher attached by single-stranded DNA, so when the target DNA is detected, Cas12a cleaves the probe, freeing the fluorophore and producing a fluorescent signal.
This technique, dubbed DETECTR, was originally developed by Jennifer Doudna, a biochemist at the University of California, Berkeley and the codeveloper of CRISPR-Cas9 genome editing, and her team in 2018, but it had not previously been tested in microgravity.16
“We wanted to test whether this technology worked in microgravity because it is such an awesome technology,” said Pines. “It has so many potential applications and it could be ideal for long space missions. They don’t need to use fancy machines or send the samples back to Earth. They can have results almost in real time.”
However, said Pines, “We don’t fully understand the effects of microgravity on biological reactions.” So, researchers designed a proof-of-concept study to test whether this technique could identify synthetic genomic sequences from three different species: a bacterium, a fungus, and an insect.
The researchers found that the test was still extremely sensitive in microgravity: it identified the target DNA in attomolar concentrations.17 In the future, researchers could use this technology to design kits to diagnose diseases in astronauts, study how the microbiome of the station and its inhabitants changes over time, or monitor the health of space farming operations on long missions.
RPA APPLICATIONSBecause RPA is rapid, portable, and runs at low temperatures it has diverse applications. | |
 Point-of-care testing for genetic and infectious diseases |  Water quality testing |
 Antibiotic resistance detection |  Agricultural pathogen monitoring |
References
- The Nobel Prize in Chemistry 1993. NobelPrize.org.
- Polymerase Chain Reaction (PCR) Fact Sheet. Genome.gov.
- Suyama Y et al. Complementary combination of multiplex high-throughput DNA sequencing for molecular phylogeny. Ecological Research. 2022;37(1):171-181.
- Piepenburg O et al. DNA Detection Using Recombination Proteins. PLoS Biol. 2006;4(7):e204.
- Silva G et al. Rapid and specific detection of Yam mosaic virus by reverse-transcription recombinase polymerase amplification. J Virol Methods. 2015;222:138-144.
- Zhao G et al. Development of a recombinase polymerase amplification combined with a lateral flow dipstick assay for rapid detection of the Mycoplasma bovis. BMC Veterinary Research. 2018;14(1):412.
- Seely S et al. Point-of-Care Molecular Test for the Detection of 14 High-Risk Genotypes of Human Papillomavirus in a Single Tube. Anal Chem. 2023;95(36):13488-13496.
- Nelson MM et al. Rapid molecular detection of macrolide resistance. BMC Infectious Diseases. 2019;19(1):144.
- Luo N et al. Establishment of methods for rapid detection of Prymnesium parvum by recombinase polymerase amplification combined with a lateral flow dipstick. Frontiers in Marine Science. 2022;9.
- Liang L et al. Development of a multi?recombinase polymerase amplification assay for rapid identification of COVID 19, influenza A and B. J Med Virol. Published online September 20, 2022:10.1002/jmv.28139.
- Cox MM. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol. 2007;8(2):127-138.
- Lobato IM, O’Sullivan CK. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Analyt Chem. 2018;98:19-35.
- Murray CJL et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet. 2022;399(10325):629-655.
- Strayer-Scherer A et al. Recombinase Polymerase Amplification Assay for Field Detection of Tomato Bacterial Spot Pathogens. Phytopathology. 2019;109(4):690-700.
- Conrad CC et al. A Sensitive and Accurate Recombinase Polymerase Amplification Assay for Detection of the Primary Bacterial Pathogens Causing Bovine Respiratory Disease. Frontiers in Veterinary Science. 2020;7.
- Chen JS et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436-439.
- Alon DM et al. CRISPR-based genetic diagnostics in microgravity. Biosens Bioelectron. 2023;237:115479.






