A rapid isothermal amplification technique enables pathogen identification and antibiotic resistance detection in low-resource settings.
Recombinase polymerase amplification (RPA) is a technique for rapidly copying segments of DNA. Unlike polymerase chain reaction (PCR), it does not require thermal cycling, so less equipment is needed.
HOW RPA WORKSResearchers require three defining elements for RPA: uvsX recombinase proteins, single-stranded binding proteins, and strand-displacing DNA polymerases. They also need primers and nucleotides to amplify the target DNA. |  |
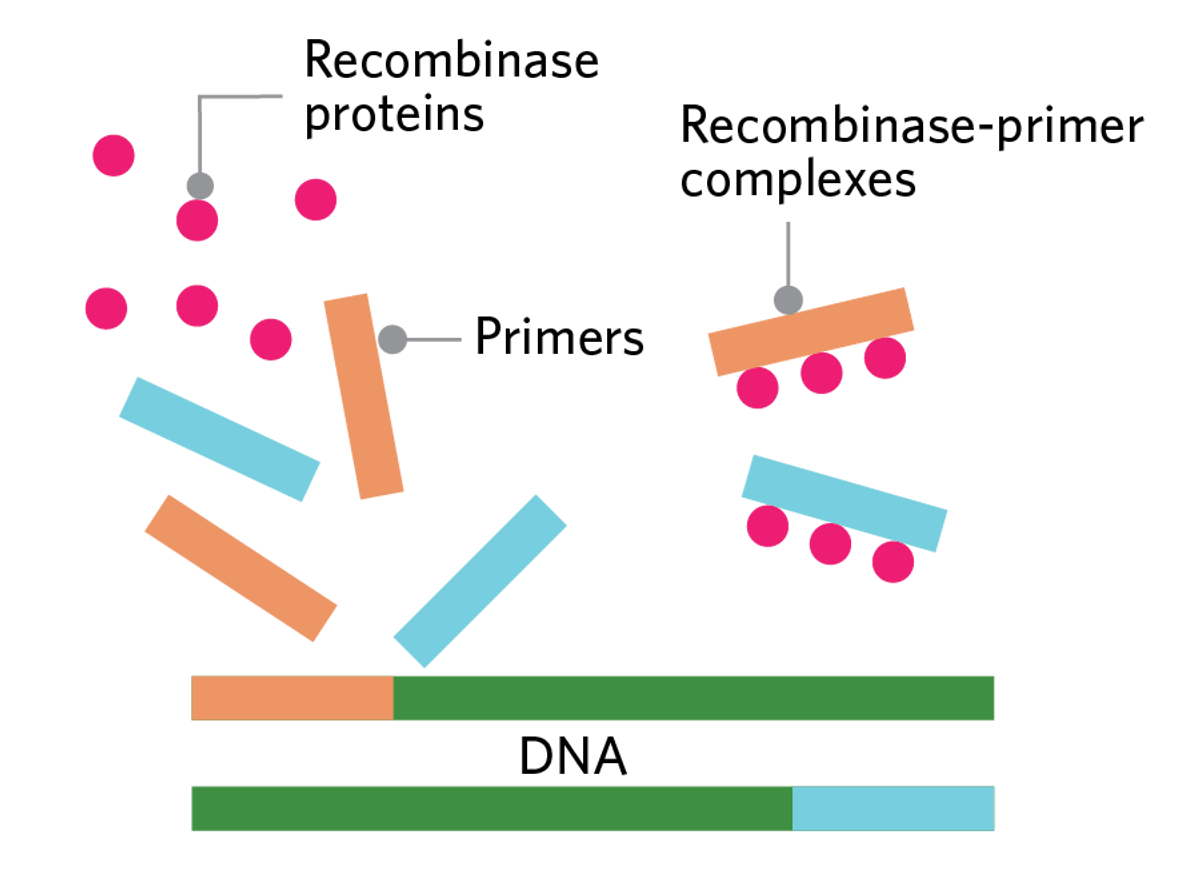 | First, the primers bind to the uvsX recombinase proteins to form recombinase-primer complexes. |
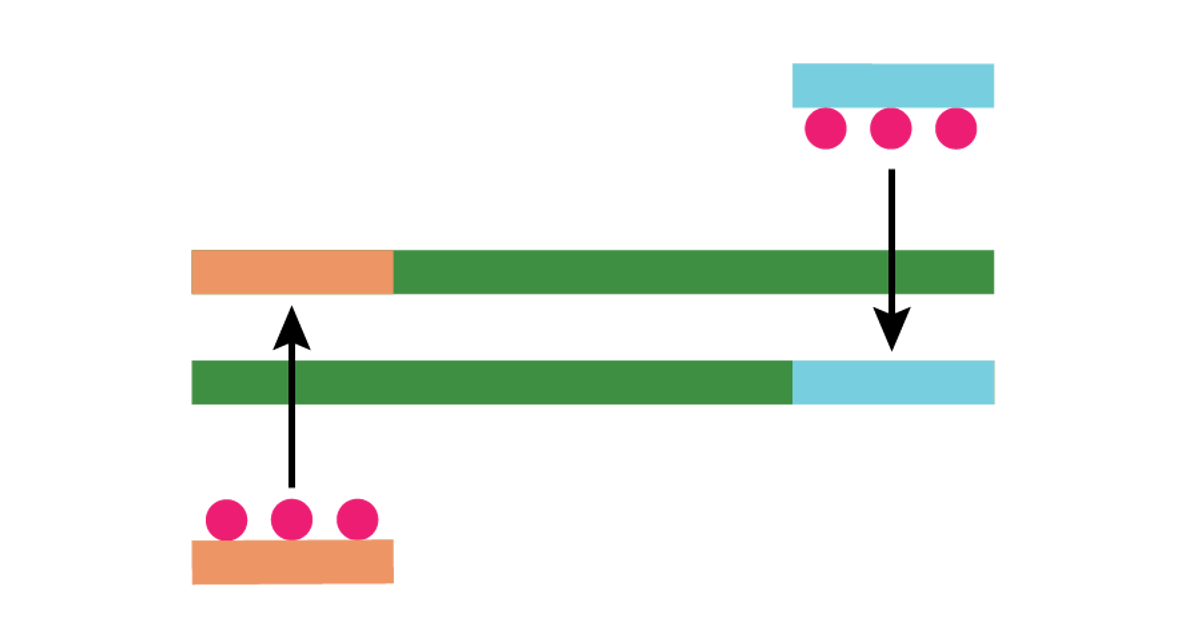 | Then the recombinase inserts the primers into complementary sites in the DNA. |
 | Single-stranded binding proteins bind to the displaced DNA strand and stabilize it. |
 | The recombinase disassembles and the strand-displacing DNA polymerase binds to the 3’ end of the primer. |
 | The polymerase elongates the primer. |
 | The target DNA strands are duplicated.1 |
RPA APPLICATIONS
Because RPA is rapid, portable, and runs at low temperatures—even body heat can be enough2—it has applications for point-of-care testing for genetics3 and infectious diseases,4 water quality testing,5 antibiotic resistance detection,6 and agricultural pathogen monitoring7.
 Point-of-care testing for genetic and infectious diseases |  Water quality testing |
 Antibiotic resistance detection |  Agricultural pathogen monitoring |
References
- Lobato IM, O’Sullivan CK. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Analyt Chem. 2018;98:19-35.
- Crannell ZA et al. Equipment-Free Incubation of Recombinase Polymerase Amplification Reactions Using Body Heat. PLOS ONE. 2014;9(11):e112146.
- Natoli ME et al. Allele-Specific Recombinase Polymerase Amplification to Detect Sickle Cell Disease in Low-Resource Settings. Anal Chem. 2021;93(11):4832-4840.
- Seely S et al. Point-of-Care Molecular Test for the Detection of 14 High-Risk Genotypes of Human Papillomavirus in a Single Tube. Anal Chem. 2023;95(36):13488-13496.
- Kunze A et al. On-Chip Isothermal Nucleic Acid Amplification on Flow-Based Chemiluminescence Microarray Analysis Platform for the Detection of Viruses and Bacteria. Anal Chem. 2016;88(1):898-905.
- Nelson MM et al. Rapid molecular detection of macrolide resistance. BMC Infectious Diseases. 2019;19(1):144.
- Silva G et al. Rapid and specific detection of Yam mosaic virus by reverse-transcription recombinase polymerase amplification. J Virol Methods. 2015;222:138-144.
Read the full story.





