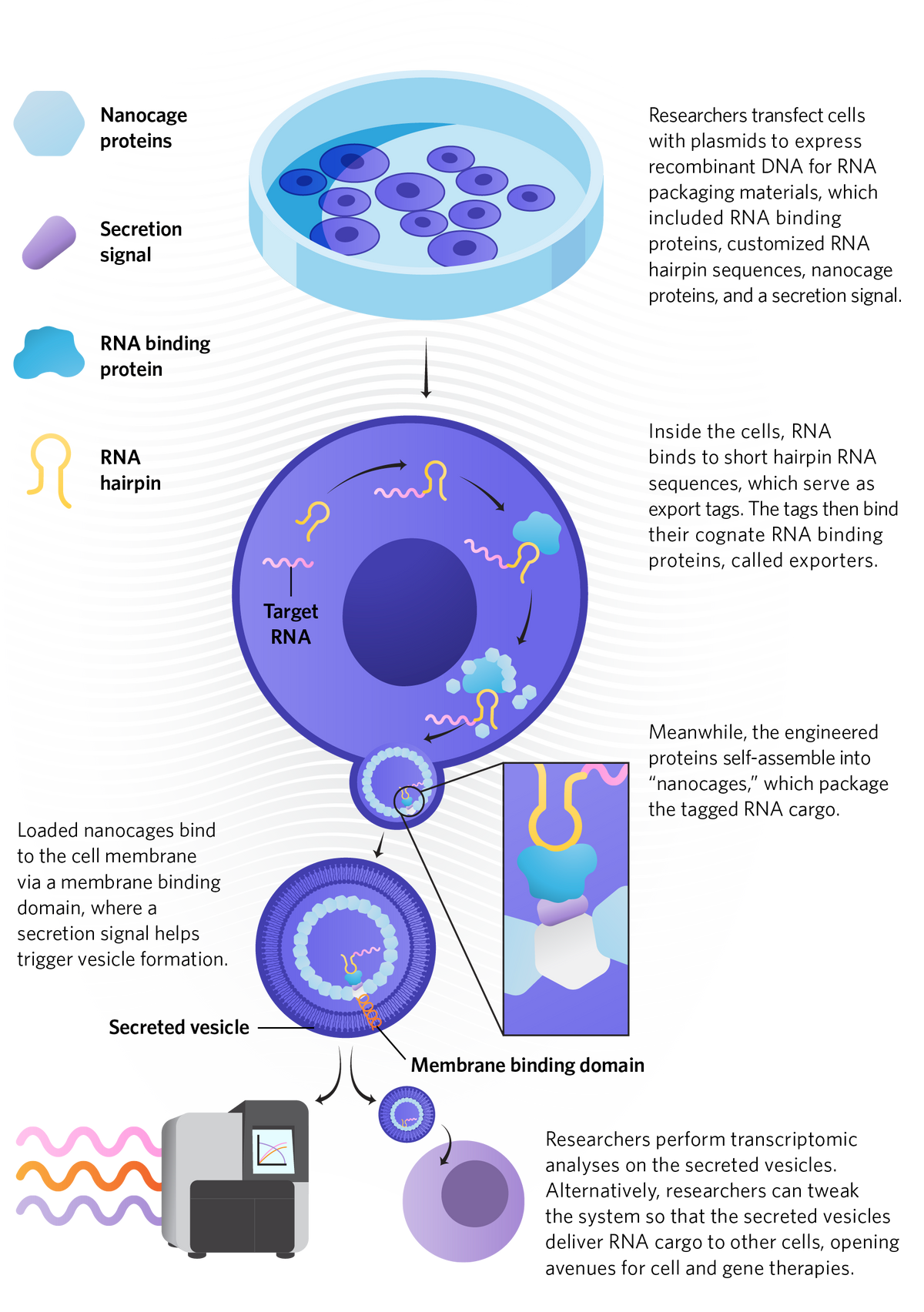
Save the Sample
Researchers must plan for the future of their lab materials long before they decide to move on.
When I was wrapping up my graduate studies, questions about the future of my project loomed large in my mind. The uncertainty around the fate of my oligonucleotide tubes felt like a cliffhanger moment in the season finale of a beloved series that has yet to be renewed for the following season. I recall the relief when a colleague took over my project, and I was spared the horror of having to pull the plug on the samples that I invested so much time and effort in.
Even though I had spent only four years on the project, it warmed my heart to know that my tubes would retain their spot in the freezer a few years longer. So, when a retiring academic recently posted on social media about the impending doom of their freezer boxes with the end of their flourishing career, I could imagine the utter disappointment of losing a lifelong work. Scrolling through the comments from other researchers, I realized that this was sadly a common occurrence.
In this age of automation and digitization, one might expect better alternatives for handling biological materials. Options such as storing them in a repository or shipping them to collaborators exist, but these solutions often require planning ahead—a luxury that not many can afford. Between helping their students graduate and wrapping up other pending projects, by the time researchers think of their samples, it is often too late. Even for the early planners, the options might be limited depending on the type of sample, particularly for those working in a niche field. Lastly, sample storage may require immaculate record keeping, and let’s face it, some researchers’ Da-Vinci-Code-level-hard-to-crack lab notebooks might not be adequate.
Perhaps with improved electronic record keeping and growing bio-storage solutions, we might not need to take up the #SavetheSample cause in the future. Until then, we would love to hear from all of you about your recommended solutions for keeping years of work from going down the drain (sometimes literally).